- Introduction
- Background and Function of Rex-1 in Mouse
- Analysis of Rex1 (Zfp42) Function in Embryonic Stem Cell Differentiation.
- Evaluation of Rex-1
- Function of Rex
- Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX
- The transcription factor YY1
- DNA-binding motifs of YY1, YY2 and REX1
- Conclusion
- References
- Footnotes
Introduction
The genetic expression of specified cell types is a clear achievement of activities of gene in the development of organisms. In this category, REX1 has the longest history as a marker gene that belongs to stem cells of undifferentiated pluripotent which dates back to 1989. The function of Rex-1 has remained mysterious ever since time. Rex 1/Zfp42 is very useful in marking undifferentiated the pluripotent state of stem cells. This paper will discuss the background, functions, expressions, and evaluation of Rex1. The connection between Rex 1 and YY1, as well as the link between Rex1 and stem cells, will be discussed adequately.
Background and Function of Rex-1 in Mouse
Rex-1 (zfp42) is classed as constituting zinc finger protein which expresses undifferentiated stem cells in embryos and adults. Through total-transformation of retinoic acid induction and murine differentiation of embryonic stem (ES) cells, the intensity of Rex-1 mRNA drops in numerous folds.
At first, the gene was recognized as a consequence of depression in occurrence upon the treatment of RA.1 Rex-1 constitutes four (4) cys-His motifs of encoded zinc-finger protein acid which is very significantly similar to the factor family of YY1 transcription found in the domain of zinc-finger. Apart from its presence in ES cells, Rex-1mRNA is present in stem cells of F9 teratocarcinoma2, as well as in statistical germ cells. The importance of Rex1 was demonstrated by the reprogramming of an adult mouse (Figure 1).

Australian Tampa scientists from MIM Research “reprogrammed” fat cells of an adult mouse and converted stem cells from neural cells differentially to pluripotency (Figure 2). They achieved this because:
‘the cells are “induced pluripotent stem cells” (iPS), (and) are nearly identical to the naturally occurring pluripotent stems cells’4
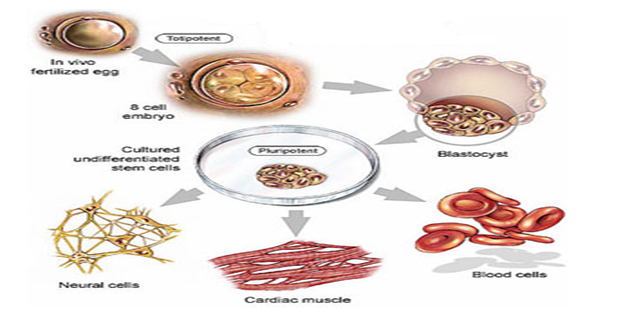
Analysis of Rex1 (Zfp42) Function in Embryonic Stem Cell Differentiation.
Embryonic Stem Cells (ES) Cells in Mouse are undifferentiated and are obtained from the internal constituent of the cells blastocyst as stated by Martin.6 ES Cells are equally pluripotent in nature; by this, the cells could be differentiated into forms that are obtained from three main germ coverings, which include: the mesoderm, the ectoderm, and the endoderm.7,8 Martin states that the differentiation of ES Cells supposes, in terms of cell culture, the existence of an intact blastocyst.9
This then infers that ES Cells are of high use as models for the investigation of embryogenesis processes and for differentiation.3 In intense consideration of ES cells, a network of the molecular transcription factors, which involves Oct 4, Sox 2, and Nanog, studies have proved adequately an accelerating system that is positive in the maintenance of self-renewal in ES cells.10
Worth note is the fact of the difference in expression for GCNF and p53 which is never repressed in the self-renewal of Nanog11-14 Studies have indicated that ES cells can be retained under standard cultural condtions15 Where there is the availability of a vitamin A metabolite called retinoic acid, ES Cells could also differentiate effectively.16 Retinoic acid has been defined as:
‘a ligand for the retinoic acid receptors (RARs), encoded by the genes RAR!, RAR”, and RAR#’.17
Normally, the activities of RA are ruled over by receptors. It has also been shown how RA arbitrates to ES Cell differentiation, partially, by circuitously attenuating the induction of LIF Stat3 activation.18 It was discovered through Laboratory analysis that Rex (zfp42) is a factor of transcription of the zinc finger. Genetic studies have also revealed that:
‘The gene was initially identified as a result of the reduction in its expression upon RA treatment’.4,19
This gene had been recognized, in the first place, resultant of a reduction that is associated with the reaction on the treatment of RA. Rex-1 constitutes proteinic zinc-finger acid which has motifs of Cys-His and is a protein that has the remarkable resemblance of the YY1 factor of transcription for domains of the zinc finger.20 Expression of protein and Rex-1mRNA was also noticed in a number of adult cell types in humans.21 Laboratory research on Rex-1 promoter is found to contain an Oct4 uniting site22. Of particular concern, Oct4 can repress or activate Rex-1 transcript; an action that relies on Oct4’s level.15 The roles of Sox2 and Nanog in transactivation of Rex-1 are found to be elucidative.23 Similarly, it was confirmed that the intensity of Rex-1mRNA dwindles to correspond with the introduction of differentiation, with or without RA’s presence.24
Presently, Rex1 is found to have broad usage as a marker of stem cells.24-27 Disorder of both Rex-1 gene alleles in teratocarcinoma F9 stem cells has been observed.26 This resulted in a generated impaired visceral endoderm differentiation.28 Related research29 agrees with the hypothesis which stipulates the vital role played by Rex-1 protein in ES Cell differentiation. To consolidate functions characterized by Rex-1, Rex-1 alleles were then disrupted in J1 murine ES Cells using two homologous rounded recombinations. Rex-1 was produced to over-express ES Cells. Gene microarray analyses were achieved to categorize Rex-1 potential target genes.
Evaluation of Rex-1
In an effort to examine the exact role played by Rex-1 in upholding pluripotency, a chain of ES cells were genetically engineered in line with attaining functional loss and gain analyses.5,30,31 In the case of functional loss analysis, endogenous alleles of Rex-1 were disrupted through targeting of predictable genes via unified conjuncture in ES cells. The result is discussed:
The knock-out (KO) allele wasa functionally null allele because the first 100 bp of the open reading frame in the exon 4 including the start codon which was replaced by the pacEGFP chimeric gene cassette containing the puromycin-resistant genePACac) and the green fluorescent protein (GFP) cDNA.30
Fascinatingly, the entire used puromycin-resistant cloned cells which were acquired through KO transfection vector achieved the desired alleles targeted.
These constitute lined Rex1+/- ES cells (RKPG9) that were cultured with great-dose of puromycin in other to get the Rex1-/- ES cell appearance produced by means of impulsive gene alteration.
Function of Rex
To typify Rex-1 function(s) more lengthily, Rex-1 was double-knocked-out in ES cell lines.6, 33 It was proposed that the activity of Rex-1 depletes the induction of retinoic acid differentially in ES cells. An enhanced Nanog expression in ES cells indicates the necessity for the diminish of its level for a vitro lineage assurance.34 Evaluation were made on Rex-1 genetic over expression; the restoration of Rex-1 was achieved in the determination Rex-1’s function.
Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX
YY1 is a factor for transcriptional DNA-binding which is present both in invertebrates and in vertebrates. Search through Databases recognized sequences linked with 62 YY1 from the numerous genome sequences that are obtainable from insects to humans.7, The sequences are typical of lofty levels of conservation of sequences in the ranges of 66% to 100% comparison35. Phylogenetic analyses reveal replication of events in YY1 for many more varying lineages, flies, fish, and mammals inclusive. Retroposition is accountable for producing a duplicate in PHOL and flies from PHO36, as well as double mammalian placental duplicates.
It is directly responsible for Rex-1 Reduced Expression from YY1. Motif studies on DNA-binding have illustrated the continual binding of YY2 to identical unified sequences as YY1, except, with a relatively lesser resemblance.33 In disparity, Rex-1 attaches to divergent motifs of DNA from YY1, except the attachment of YY1 and Rex-1 which shares a number of core-region (5’-CCAT-3’) similarities.37 This connotes the duplicates of Rex-1 and YY2, even though they are produced through comparable retroposional events which have gone through numerous selected schemes to adhere to emergent mammalian placental role.38-40
Largely, the preservation of REX1 and YY2 in all mammals with placental suggest strongly the co-evolution of each duplicate which has unique eutherian features of mammals.
The transcription factor YY1
The Transcription Factor YY1 (otherwise known as Yin Yang 1) has been defined by as:
Gli–Kruppel type zinc finger protein, and can function as a repressor, activator or transcription initiator depending upon the sequence context of YY1-binding sites with respect to other regulator elements (1)’.41
The protein constitutes a domain of DNA-binding is tracked at the C-terminus as well as a number of similar adoptive domains which may exist at N-terminus and display activation, protein or protein coordinated actions, as well as repression.8,42 YY1 acts together with a number of other major transcription factors that may include TAFs, TBP, SP1 and TFIIB, and also may include histone-regulating complexes like PRMT1, p300,Polycomb, and p300 complexes.
Several other viral and cellular genes are checked and put in place by YY1. Studies have put it that at least 10% of genes in mice constitute sites for YY1 binding close to their regions of promotion. A further set of researches has put it that imprinted genes of mice have quite strange YY1-binding sites arranged in tandem arrays which exist in locations for control, thus expressing genomic imprint roles in mammals. Several experiments on mouse mutagenesis revealed the vital position of dependence of dosage on YY1 in the course of development and organization of cell cycle.43 The evolution of YY1 is properly protected in all lineages of invertebrates and in vertebrates.
It is shown in a number of species of mouse44, and even in flies with comparable genes as the YY1, Pho-like (or PHOL), and Pleiohomeotic9,. Foundamentally:
‘PHO is one of the DNA-targeting proteins for the Polycomb complex and the phenotypes of pho-deficient mutants can be rescued by mammalian YY1’.45
In mouse’s genomes, a further two genes associated with YY1 are found. These are Reduced Expression 1 (REX1) and Yin Yang 2 (YY2). YY2’s function can be linked to that of YY146, and is YY1 replicate due to the intronless structure it has and intron’s position relative to a different genes of X-chromosomes, Mbtps2.46 Another study says:
Rex-1 was independently discovered, before the identification of YY1, due to its unique expression profile: dramatic decline of expression after retinoic acid-induced differentiation of F9 murine teratocarcinoma stem cells.47
Subsequently, studies on Rex-1 are based on marker of stem cells which are Oct3/4 controlled.10,47 Comparative studies by stressed that Rex-1 is subfamily to the YY1 subfamily.
DNA-binding motifs of YY1, YY2 and REX1
Investigation on the function of outcomes of a number of chosen zinc-finger-domain imposed pressures on Rex-1, YY1 and YY2 were distinguished by self DNA-binding motifs. In a particular experiment, domains of zinc-finger of certain proteins was sub-cloned in lower segments of GST protein, which constitute fushed proteins in bacterial. This are agarose-bead fixed and were granted the freedom of duplex DNA binding which was obtained from haphazard oligonucleotide sequence (4n=15).48

In the situation of YY1, twenty out of thirty-four DNA bounds had DNA motifs which either had a ideal matching or a base deprivation from the regular YY1 sequence of consensus. The rest 14 DNA bounds continue showing similarities as YY1, except they had a middling base 2 difference from YY1.11 This illustrates the central position which YY1 has in Zinc-finger domain. The analysis is as expressed:
The sequences of DNAs bound by YY1 (left), YY2 (middle) and REX1 (right) are shown with the clone numbers on the right. The uppercase sequences are derived from the randomized portion of the input DNAs for binding whereas the lowercase dinucleotides represent the surrounding, fixed portion of the input DNAs. The majority of the DNAs bound by both YY1 and YY2 contain the known YY1 consensus motif (CGCCAT.TT), which is marked blue in the forward direction and by red in the reverse direction.49
The Binding motifs of DNA on YY1 and Rex-1 is performed with the use of shift gel assays. In YY1’s situation, a number of duplex oligonucleotides are used as identified in the demonstration of subtle YY1 properties. Very expectedly, the domain of DNA-binding, as a GST-YY1 fussional part, expressed a nearly same DNA partern as that of YY1 protein. This is expressed as follows:
The GST-YY1 protein is methylation-sensitive: methylation on the upper strand is inhibitory to the binding. One base change in this CpG site, either CpA or TpG, somewhat reduced the affinity of the YY1 binding, but still allowed YY1 binding to these probes. The DNA-binding domain of YY2 also showed a similar pattern of DNA binding: methylation-sensitive binding and subtle effects by single base changes caused by the CpG site.
However, the DNA-binding affinity of YY2 is much weaker than YY1 based on the results derived from our control experiments for gel shift assays (Supplementary Data 4). We have also tested some of the DNAs that contain two motifs within the randomized portion of the target DNAs. We did not observe any difference in binding between the duplex DNAs with two binding motifs versus single binding motif (data not shown). Overall, the DNA-binding patterns of YY1 and YY2 appear to be similar except for the fact that the binding affinity of YY2 is much weaker than YY1, consistent with the observed relaxation of evolutionary constraint on the DNA-binding domain of YY2.50
Conclusion
This paper discusses the relevance of Rex-1 to genetic ‘reprogramming’12, especially in mouse, and expresses the usefulness it has in marking undifferentiated pluripotent state of stem cells.51 This is achieved through discussing background, functions, expressions, and evaluation of Rex1. And then, the connection between Rex 1 and YY1, as well as link between Rex1 and stem cells is discussed adequately.
References
Bestor A & Bourc’his D, Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb,. Quant, London, 2004.
Betsy A, Melissa R, Christine K & Gudas J, An Octamer Motif Contributes to the Expression of the Retinoic Acid-Regulated Zinc Finger Gene Rex-1 (Z-42) in F9 Teratocarcinoma Cells, MOLECULAR AND CELLULAR BIOLOGY 1993: 2919- 2928.
Bos J, Lee R, Cruz-Munoz W, Bjarnason GA, Christensen JG & Kerbel RS, Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis, Cancer Cell. 2009;15:232–9*T
Carmeliet P & JainI R, Nature. 2000;407:249–57.
Das B, Tsuchida R , Malkin D, Koren G, Baruchel S, Yeger H, Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells, 2008;26:1818–30.
David C, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Müller-Höcker J, Kitajima S, Lickert H, Rupp R & Franz W, MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling, Nature Cell Biology, vol 10, 2008, pp. 338 – 345.
David R, Brenner C, Stieber J, Schwarz J, Brunner S, Vollmer M, Mentele E, J Müller-Höcker, S Kitajima, H Lickert, R Rupp & W Franz, MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling, Nature Cell Biology, vol. 10, 2008, pp. 338 – 345.
Division of Molecular Biology and Cell Engineering, Department of Regenerative Medicine, Research Institute, International Medical Center of Japan, 1-21-1 Toyama, Shinjyuku-ku, Tokyo 162-8655, Japan, 2010.
Gassmann M, Wartenberg M, Mcclanahan T, Fandrey J, Bichet S, Kreuter R, Acker H & Bauer C, Differentiating embryonic stem cells as an in vitro model of early erythropoiesis, Vitro vol. 9, issue 4, August 1995, pp. 429-433.
Geza S & Robert P, Characterization ofthe mouse gene that encodes the6/YY1/NF- El/UCRBP transcription factor, Biochemistry Vol. 90, 1993, pp.5559- 5563.
Gordan JD, Bertout JA, Hu CJ, Diehl JA & Simon MC, HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity, Cancer Cell. 2007;
Huang S, Laoukili J, Epping M, Koster M, Hölzel M, Westerman B, Nijkamp W, A Hata, S Asgharzadeh, R Seeger, R Versteeg, R Beijersbergen, & R Bernards, ZNF423 Is Critically Required for Retinoic Acid-Induced Differentiation and Is a Marker of Neuroblastoma Outcome, Cancer Cell, vol. 15, issue 4, 2009, pp. 328–340.
Jain R, di Tomaso E, Duda D, Loeffler J, Sorensen AG & Batchelor TT, Angiogenesis in brain tumours, Nat Rev Neurosci. 2007;8:610–22.
John M, Li M, Anita B & Jeremy N, The Hypoxic Microenvironment Maintains Glioblastoma Stem Cells and Promotes Reprogramming towards a Cancer Stem Cell Phenotype, Cell Cycle, 2009; 8(20): 3274–3284.
Kiel M & Morrison J, Uncertainty in the niches that maintain haematopoietic stem cells, Nat Rev Immunol. 2008;8:290–301.
kim J, Faulk C & Kim J, Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX, Nucleic Acids research, vol. 35, issue, 10, 2010, pp.3442-3452.
Kim J, Hinz A, Bergmann A, Thompson M, Ovcharenko I, Stubbs L & Kim, Identification of clustered YY1 binding sites in imprinting control regions, Genome Res, vol. 16, 2006, pp.901-911.
Li P, Tong C, Mehrian-Shai C, Jia C, Wu N, Yan Y, Maxson R, Schulze E, Song H, Hsieh C, Pera M & Ying Q, Germline Competent Embryonic Stem Cells Derived from Rat Blastocysts, Cell, vol. 135, issue 7, 2008, pp. 1299- 1310.
Liang J, Hung D, Schreiber S & Clardy J, Structure of the human 25 kDa FK506 binding protein complexed with rapamycin, J.Am.Chem.Soc., vol. 118, 1996, pp. 1231-1232.
Liu R, Wang X, Chen G, Dalerba P, Gurney R, Hoey T, et al, The prognostic role of a gene signature from tumorigenic breast-cancer cells, N Engl J Med. 2007; 356:217–26
Mani S, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al, The epithelial- mesenchymal transition generates cells with properties of stem cells, Cell. 2008, pp.704–15.
Martin H, Human ES Cells and a Blastocyst from One Embryo: Exciting Science but Conflicting Ethics?, Cell Stem Cell, vol. 2, issue 2, 2008, pp. 103- 104.
Monash Institute of Medical Research, A reprogrammed adult mouse, Australia, 2010.
Onnet D & Dick J, Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell, Nature Med, vol. 3, 1997, pp.730–737.
Pan G & Pei DQ, Identification of two distinct transactivation domains in the pluripotency sustaining factor nanog, Cell Res, vol. 13,2003, pp.499–502.
Pereira L, Yi P & Merrill B, Repression of Nanog Gene Transcription by Tcf3 Limits Embryonic Stem Cell Self-Renewal, Mol Cell Biol, vol.26, issue 20, 2006, pp. 7479–7491.
Pereira L, Yi F, & Merrill B, Repression of Nanog Gene Transcription by Tcf3 Limits Embryonic Stem Cell Self-Renewal, Mol Cell Biol, vol. 26, issue 20, 2006, pp. 7479–7491.
Pistollato F, Chen HL, Rood BR, Zhang HZ, D’Avella D, Denaro L, et al, Hypoxia and HIF1alpha repress the differentiative effects of BMPs in high-grade glioma, Stem Cells,vol. 27, 2009, pp.7–17.
Reya T, Morrison S, Clarke F & Weissman L, Stem cells, cancer, and cancer stem cells, Nature,vol.414, issue 105, 2001, pp.105.
Shi Y, Seto E, Chang L & Shenk T, Transcriptional repression by YY1, a human GLI- Kruppel-related protein, and relief of repression by adenovirus E1A protein, Cell, vol 67, issue 2, 1991, pp.377–88.
Singh S, Clarke I, M Terasaki, Bonn V, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res, vol. 63, 2003, pp.5821–8.
Siomi H & Dreyfuss G, A nuclear localization domain in the hnRNP A1 protein, J Cell Biol, vol. 129, 1995, pp.551–560.
Slaughter J, Parker J, Martens M, Smarr M & Hewett J, Clinical Outcomes Following a Trial of Sertraline in Rheumatoid Arthritis, Psychosomatics 2002 43:36-41.
Smith A, Brownawell A, Macara I G. Nuclear import of Ran is mediated by the transport factor NTF2, Curr Biol, vol. 8, 1998, pp.1403–1406.
Stambler B, Laurita K, Shroff S, Hoeker G & Martovitz N, Heart Rhythm, vol. 6, issue 6, 2009, pp. 776-783.
Stein G, T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro, J Cell, Adibaba, 1979.
Stem Cell Innovations Produces Human Pluripotent Stem Cells, Hyscience, 2006.
Stem cells from Bench to Bedside, J Exp Med, vol. 178, issue 3, 1993, pp.1085–1090.
Stevens L, The development of transplantable teratocarcinomas from intratesticular grafts of pre- and postimplantation mouse embryos, Dev. Biol, 1970, 21:364-382.
Sui G, Affar S, Shi Y, Brignone C, Wall N, Yin P, Donohoe M, Luke M, Calvo D et a, Yin Yang 1 is a negative regulator of p53. Cell 2004; 117:859-872.
Vescovi AL, Galli R & Reynolds BA, Brain tumour stem cells, Nat Rev Cancer, vol. 6, 2006, pp.425–36.
Yaseen N R, Blobel G. Cloning and characterization of human karyopherin beta-3. Proc Natl Acad Sci USA, vol.99, 1997, pp.4451–4456.
Yuan Y, Corbi N, Basilico C & Dailey L, Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3, Genes Dev, vol. 9, 2003, pp.2635-2645.
Zhang D, Vuocolo S, Masciullo V, Sava T, Giordano A , Robert D & Soprano J, Cell cycle genes as targets of retinoid induced ovarian tumor cell growth suppression, Oncogene, vol. 20, Numb. 55, pp.7935-7944.
Zhu W, Lossie A, Camper S & Gumucio G, Chromosomal localization of the transcription factor YY1 in the mouse and human, Mamm Genome, vol. 5, issue 1994, pp 234–6.
Footnotes
- J R Slaughter, JC Parker, MP Martens, KL Smarr & JE Hewett, Clinical Outcomes Following a Trial of Sertraline in Rheumatoid Arthritis, Psychosomatics 2002 43:36-41.
- M. Gassmann, M Wartenberg, T Mcclanahan, J Fandrey, S Bichet, R Kreuter, H Acker & C Bauer, Differentiating embryonic stem cells as an in vitro model of early erythropoiesis, Vitro vol. 9, issue 4, 1995, pp. 429-433.
- Monash Institute of Medical Research, A reprogrammed adult mouse, Australia, 2010.
- A Betsy, R Melissa, K Christine & J Gudas, An Octamer Motif Contributes to the Expression of the Retinoic Acid-Regulated Zinc Finger Gene Rex-1 (Z-42) in F9 Teratocarcinoma Cells, MOLECULAR AND CELLULAR BIOLOGY 1993: 2919-2928.
- Stem Cell Innovations Produces Human Pluripotent Stem Cells , Hyscience, 2006.
- 3H Martin, Human ES Cells and a Blastocyst from One Embryo: Exciting Science but Conflicting Ethics?, Cell Stem Cell, vol. 2, issue 2, 2008, pp. 103-104.
- M John, Z Li, B Anita & N Jeremy, The Hypoxic Microenvironment Maintains Glioblastoma Stem Cells and Promotes Reprogramming towards a Cancer Stem Cell Phenotype, Cell Cycle, 2009; 8(20): 3274–3284.
- Stem cells from Bench to Bedside, J Exp Med, vol. 178, issue 3, 1993, pp.1085–1090.
- S Geza & P Robert, Characterization ofthe mouse gene that encodes the6/YY1/NF-El/UCRBP transcription factor, Biochemistry Vol. 90, 1993, pp.5559-5563.
- L Boyer, T Lee, M Cole, S Johnstone, S Levine, J Zucker, M Guenther, R Kumar, H Murray, R Jenner, D Gifford, D Melton, R Jaenisch, & R Young, Core transcriptional regulatory circuitry in human embryonic stem cells, Cell, vol. 122, 2005, pp. 947-956.
- 4P Gu, D LeMenuet, A. C. Chung, M. Mancini, D. A. Wheeler, and A. J. Cooney. 2005. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell. Biol. 25:8507-8519.
- T Lin, C Chao, S Saito, S. J. Mazur, M. E. Murphy, E. Appella, & Y. Xu, induces differentiation of mouse embryonic stem cells by suppressing Nanog expression, Nat. Cell Biol, vol. 7, 2005, Pp.165-171.
- C David, C Brenner, J Stieber, F Schwarz, S Brunner, M Vollmer, E Mentele, J Müller-Höcker, S Kitajima, H Lickert, R Rupp & W Franz, MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling, Nature Cell Biology, vol 10, 2008, pp. 338 – 345.
- D Onnet & J Dick, Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell, Nature Med, vol. 3, 1997, pp.730–737.
- Merrill, B. J., H. A. Pasolli, L. Polak, M. Rendl, M. J. Garcia-Garcia, K. V. Anderson, and E. Fuchs. 2004. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development 131:263-274.
- L Pereira, F Yi & B Merrill, Repression of Nanog Gene Transcription by Tcf3 Limits Embryonic Stem Cell Self-Renewal, Mol Cell Biol, vol.26, issue 20, 2006, pp. 7479–7491.
- B Stambler, K Laurita, S Shroff, G Hoeker & N Martovitz, Heart Rhythm, vol. 6, issue 6, 2009, pp. 776-783.
- Christof Nolte and Robb Krumlauf D Zhang, S Vuocolo, V Masciullo, T Sava, A Giordano, D Robert & J Soprano, Cell cycle genes as targets of retinoid induced ovarian tumor cell growth suppression, Oncogene, vol. 20, Numb. 55, pp.7935-7944.
- P Li, C Tong, R Mehrian-Shai, L Jia4, N Wu, Y Yan, R Maxson, E Schulze, H Song, C Hsieh, M Pera, & Q Ying, Germline Competent Embryonic Stem Cells Derived from Rat Blastocysts, Cell, vol. 135, issue 7, 2008, pp. 1299-1310.
- SK Singh, ID Clarke, M Terasaki, VE Bonn, C Hawkins, J Squire, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res, vol. 63, 2003, pp.5821–8.
- R David, C Brenner, J Stieber, F Schwarz, S Brunner, M Vollmer, E Mentele, J Müller-Höcker, S Kitajima, H Lickert, R Rupp & W Franz, MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling, Nature Cell Biology, vol. 10, 2008, pp. 338 – 345.
- L Pereira, F Yi, & B Merrill, Repression of Nanog Gene Transcription by Tcf3 Limits Embryonic Stem Cell Self-Renewal, Mol Cell Biol, vol. 26, issue 20, 2006, pp. 7479–7491.
- Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–36.
- S Huang, J Laoukili, M Epping, J Koster, M Hölzel, B Westerman, W Nijkamp, A Hata,S Asgharzadeh, R Seeger, R Versteeg, R Beijersbergen, & R Bernards, ZNF423 Is Critically Required for Retinoic Acid-Induced Differentiation and Is a Marker of Neuroblastoma Outcome, Cancer Cell, vol. 15, issue 4, 2009,pp. 328–340.
- M Kiel & J Morrison, Uncertainty in the niches that maintain haematopoietic stem cells, Nat Rev Immunol. 2008;8:290–301.
- P Carmeliet , RK JainI. Nature. 2000;407:249–57.
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15.
- W Zhu, A Lossie, S Camper, D Gumucio, Chromosomal localization of the transcription factor YY1 in the mouse and human, Mamm Genome, vol. 5, issue 4, 1994, pp 234–6.
- Division of Molecular Biology and Cell Engineering, Department of Regenerative Medicine, Research Institute, International Medical Center of Japan, 1-21-1 Toyama, Shinjyuku-ku, Tokyo 162-8655, Japan, 2010.
- Pan GJ, Pei DQ. Identification of two distinct transactivation domains in the pluripotency sustaining factor nanog. Cell Res. 2003;13:499–502.
- R Liu, X Wang, G Chen, P Dalerba, A Gurney, T Hoey, et al, The prognostic role of a gene signature from tumorigenic breast-cancer cells, N Engl J Med. 2007;356:217–26
- B Das, R Tsuchida, D Malkin, G Koren, S Baruchel, H Yeger, Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells, 2008;26:1818–30.
- I Chambers, D Colby, M Robertson, J Nichols, S Lee, S Tweedie, & A Smith, Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cell, Cell, vol. 113, 2003, pp. 643-655.
- Pistollato F, Chen HL, Rood BR, Zhang HZ, D’Avella D, Denaro L, et al, Hypoxia and HIF1alpha repress the differentiative effects of BMPs in high-grade glioma, Stem Cells. 2009;27:7–17.
- L Stevens, The development of transplantable teratocarcinomas from intratesticular grafts of pre- and postimplantation mouse embryos, Dev. Biol, 1970, 21:364-382.
- J Bos, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–9*T Bestor & D Bourc’his, Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb,. Quant, London, 2004.
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–47.
- G Stein, T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro, J Cell, Adibaba, 1979.
- J Kim, C Faulk, & J Kim, Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1, Nucleic Acids Res, vol.35, issue 10, 2007, pp. 3442–3452.
- H Yuan, N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635-2645.
- G Sui, S Affar, Y Shi,C Brignone, N Wall, PYin P, M Donohoe , M Luke, D Calvo et a, Yin Yang 1 is a negative regulator of p53. Cell 2004;117:859-872.
- Smith A, Brownawell A, Macara I G. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406.
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560.
- Yaseen N R, Blobel G. Cloning and characterization of human karyopherin beta-3. Proc Natl Acad Sci USA. 1997;94:4451–4456.
- Pan GJ, Pei DQ. Identification of two distinct transactivation domains in the pluripotency sustaining factor nanog. Cell Res. 2003;13:499–502
- J Kim, C Faulk, & J Kim, Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1, Nucleic Acids Res, vol.35, issue 10, 2007, pp. 3442–3452.
- Y Shi, E Seto, L Chang, T Shenk , Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein, Cell, vol 67, issue 2, 1991, pp.377–88.
- J Kim, C Faulk, & J Kim, Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1, Nucleic Acids Res, vol.35, issue 10, 2007, pp. 3442–3452.
- J kim, C Faulk & J Kim, Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX, Nucleic Acids research, vol. 35, issue, 10, 2010, pp.3442-3452
- J Kim, AHinz, A Bergmann, M Thompson, I Ovcharenko, L Stubbs, J Kim, Identification of clustered YY1 binding sites in imprinting control regions, Genome Res, vol. 16, 2006, pp.901-911.