Pharmacogenetics is a field of study that entails examining the impact of genes on the body’s response to specific medications. This field is also referred to as pharmacogenomics. Genes are made up of DNA sequences that hold information about unique physical traits. However, they also influence the safety and efficacy of drugs (Picard et al., 2017). For these reasons, a particular drug at a specified dose may elicit diverse upshots in different people, including side effects to the medication, which may be present in some people and absent in others.
Pharmacogenetic testing examines specific genes to provide information about the type of drugs and correct dosages for different people. For instance, it is estimated that people of Asian origin are ten times more likely to develop a condition known as Stevens-Johnson syndrome, which is an adverse reaction to specific drugs or infections (Nguyen et al., 2015). The HLA allele B*1502 is a biomarker that is associated with the development of this condition.
Therefore, before administering the antiepileptic drug carbamazepine, it is necessary to conduct HLA-B*1502 testing in Asian populations as a precautionary measure (Miliszewski, Kirchhof, Sikora, Papp, & Dutz, 2016). In contrast, patients with the human immunodeficiency virus with the HLA-B*5701 allele have been shown to have hypersensitivity reactions to abacavir-containing medications (Carolino et al., 2017).
Pharmacogenetic testing may be performed for one of three reasons: to determine whether a given drug will be effective, to predict the possibility of adverse side effects, and to find the appropriate dosage. Consequently, such tests may be done before starting a new medication or when current medications are not yielding the expected outcomes. However, pharmacogenetic testing can only be done for a few drugs, particularly those whose metabolism is mediated by cytochrome P-450 enzymes (Qiao et al., 2016).
Major depressive disorder (depression) is a common mental health disorder with a large burden of disease globally. Currently, many effective antidepressants are available to manage the condition. However, there is a poor response to first-line antidepressants in more than half of patients diagnosed with depression (Cipriani et al., 2018). Furthermore, most patients on antidepressants report adverse side effects. Nevertheless, in the past few decades, there have been groundbreaking discoveries concerning the effect of genetic variability on clinical attributes such as drug metabolism. This knowledge can be used to tailor treatment to individual patients (Fan & Bousman, 2019).
In it has been shown that genetic variation in cytochrome P450 (CYP450) enzymes, for instance, CYP2D6, CYP2C19, CYP2C9, and CYP3A4/5 are responsible for discrepancies in the metabolism of many pharmacological agents, including commonly used antidepressants (Altar et al., 2015). The purpose of this paper is to propose a study to conduct pharmacogenetic testing for antidepressants in a population of patients diagnosed with major depressive disorder.
Problem Statement
The incidence of adverse drug events is reported to range from 0.17% to 65%, which has increased healthcare costs in the US to approximately $76.558 billion (Meier et al., 2015). Furthermore, vast financial resources are wasted on trial and error treatments when known pharmacotherapeutic agents fail to treat medical and mental health conditions. Major depressive disorder is a rampant mental health problem whose management has not been attained optimally due to unresponsiveness to medications and nonadherence attributed to adverse drug reactions.
Pharmacogenetics entails utilizing knowledge about an individual’s genetic constitution together with additional clinical data to develop a customized medication regimen with improved efficiency and safety. Pharmacogenomic investigations have been done on numerous drugs to back apposite dosing or selection. Furthermore, pharmacogenetic evaluations are currently conducted as a procedure in the course of drug development.
Even though pharmacogenomic testing is a possible solution to the effective management of major depressive disorders, it is still underutilized in clinical practice. Possible causes for its underutilization include the lack of conviction about its impact on overall patient outcomes in major depressive disorder. Therefore, there is a need to conduct clinical trials to confirm the impact of pharmacogenomic testing on patient outcomes in depression, which is the focus of this study.
The Theoretical Framework
This study will be based on the ecological model of health. The ecological outlook is a valuable framework for comprehending the myriad of factors that affect health and wellbeing (Sallis, Owen, & Fisher, 2015). This model helps by providing a clear picture of the factors that influence precise health behaviors, for example, the social determinants of health. Consequently, ecological frameworks can be used to incorporate aspects of other theories, thereby leading to the design of an all-inclusive health promotion or disease preclusion approach.
The ecological model of health assumes that health conduct is influenced by several factors, including individual, interpersonal, institutional, community, and public factors. Individual factors, which are also known as intrapersonal factors, affect the knowledge, beliefs, attitudes, and personality of a person. Interpersonal factors encompass dealings with other people that may have positive upshots such as offering social support or negative outcomes such as hindering social growth attributed to healthy behavior. In contrast, institutional and organizational factors take into account rules, guidelines, dogmas, and informal constructions that limit or endorse healthy behaviors. Community factors consist of social norms that prevail among individuals or groups and can promote or constrict healthy conduct.
Finally, public policy factors comprise policies and laws that control health practices such as prompt diagnosis and management, which strive to prevent morbidity and mortality associated with disease (Sallis, Owen, & Fisher, 2015).
In this study, the pharmacological treatment of diseases depends on several factors (ecologies), including drug dosage, known efficacy, age, and gender of the patient as well as individual response to treatment, which is a component of the person’s genetic makeup. By elucidating the genetic factors that determine a patient’s reaction to treatment, it will be possible to control for other factors that influence response to pharmacological intervention thus ensuring optimal patient outcomes. Therefore, factors that affect the effectiveness of pharmacological treatment will be considered as the “ecological” factors in the theoretical framework.
The Purpose of the Study
The purpose of this study is to look into the clinical outcomes of the personalization of antidepressants in major depressive disorder. Genetic testing will be done to generate pharmacogenetic profiles of the participants. The key factors that will be examined include response to treatment and the incidence of adverse drug events. The first specific objective is to determine the disease remission rates when antidepressants are administered based on the drug metabolism profiles obtained by genetic testing. The second specific objective is to assess the incidence of adverse drug events when antidepressants are prescribed based on each patient’s pharmacogenetic profile.
Significance of the Study
Pharmacogenetic testing is a crucial constituent of personalized medicine that contributes towards the safe and effective prescription of drugs to patients. This process exploits the data given by a patient’s distinct genetic composition to foretell pharmacokinetic silhouettes and pharmacodynamic responses for many xenobiotics. It has been shown that pharmacogenomic testing lowers the incidence of adverse drug events, enhances response to medications and lowers the cost of health care (Hughes, 2018). Even though tremendous progress has been made in this field in the last few years, certain obstacles get in the way of maximum utilization of pharmacogenomics.
For instance, the fiscal and analytical considerations as well as the lack of conviction about the importance of pharmacogenomics among patients and clinicians. Therefore, there is a need to conduct this study to verify the benefits of pharmacogenetic testing in the treatment of major depressive symptoms.
Research Questions and Hypotheses
From the preliminary information regarding the benefits of pharmacogenetic testing, it is hypothesized that there will be higher rates in symptom remission when antidepressants are administered based on the drug metabolism profiles of patients. It is also hypothesized that pharmacogenetic testing will lead to fewer cases of adverse drug events in patients. Therefore, the two research questions related to these hypotheses are “What is the impact of pharmacogenetic testing on symptom remission of when antidepressants are used in patients with depression?” and “What is the effect of pharmacogenetic testing for antidepressants on the incidence of adverse drug events in patients with depression?”
Summary
Pharmacogenomic testing is a useful tool that helps in the customization of drug treatment. It takes advantage of a patient’s genetic composition to develop a unique drug profile and use it to choose the most appropriate medication and dosing. This process is important in the use of antidepressants to manage major depressive disorder. The purpose of this study is to determine the effect of pharmacogenomic testing on symptom remission and the incidence of adverse drug events in patients with major depressive disorder.
Method
Design
The study will follow a case-control design. This is a form of observational study where two different groups with different attributes are identified and compared based on an assumed causal attribute (Belbasis & Bellou, 2018). It is also referred to as a case-referent study. The two attributes are delineated at the beginning of the study. Based on the observed differences between the two groups, the investigator then looks back to evaluate whether there is a statistically significant difference.
Participants
The sample for this study will comprise 42 patients receiving mental health care at a healthcare facility between December 2019 and August 2020. A similar proportion of men and women will be included in the study. The mean age of the participants will be ascertained following recruitment.
Population
Participants will be drawn from a mixed population without special consideration of ethnic or racial background. Therefore, it is expected that partakers of the study will come from ethnicities such as Caucasian, African American, Asian, and Hispanic based on the ethnic composition of the population in the study site. However, all patients will have a history of major depressive disorder.
Selection Criteria
All patients will be diagnosed with depression. However, to be included in the study, the subjects need to have a score of 8 or greater on the Hamilton Depression Rating Scale (HDRS) at their initial visit to the healthcare facility. The participants will be 18 years and older and demonstrate a willingness to participate in the study. Exclusion criteria will include refusal to take part, known physical comorbidities, pregnancy, and HDRS scores lower than 8. Furthermore, patients who will be unable to undergo psycho-affective symptomatology evaluation for reasons such as serious cognitive deterioration, a deficit of awareness, compromised communication skills, and reluctance to go through a psychological test will be excluded.
Sampling Approach
Purposive sampling will be used to select the study participants. This sampling approach is also referred to as selective, judgmental, or subjective sampling. It is a form of non-probability sampling methods where the attributes of the units to be studied are determined by the investigator (Etikan, Musa, & Alkassim, 2016). In most cases, smaller sample sizes than those used in probability sampling are selected in purposive sampling.
Purposive sampling does not strive to use the study outcomes to make generalizations or statistical inferences to the population. Instead, it narrows down on specific attributes in the study population that will facilitate the answering of research questions. Therefore, the units under investigation are not representative of the population. Homogeneous purposive sampling, where units are chosen based on similar physiognomies, will be applied for this research. These features are of interest to the overall goal of the study and have been captured in the inclusion and exclusion criteria.
Recruitment
The first step of the procedure will involve informing each individual about the likelihood of using their pharmacogenetic and clinical information as part of the clinic’s research project. Prospective participants will be informed about the basics of the study, possible benefits, and shortcomings after which their authorization will be requested by having them sign informed consent forms. Each patient will then be subjected to a brief clinical interview to collect additional sociodemographic data and drug use patterns. The participants will be assured about the privacy and confidentiality of their data. Ethical clearance will be obtained from the author’s learning institution before commencing the investigation.
Participant Compensation
The study will involve the testing of pharmacological interventions for depression in the presence and absence of pharmacogenetic testing in patients receiving treatment at a health facility. Therefore, there will be no requirement for participant compensation. However, the patients will benefit from scheduled continuous monitoring of their therapeutic progress throughout the study period as deemed necessary by the principal investigator without requiring them to pay.
Power
A repeated-measures ANOVA will be used for data analysis. The number of participants in the study was determined by conducting a priori power analysis in G*Power. The expected power was 0.9 with a medium effect size of 0.5. An alpha level of 0.05 was chosen. The sample size required to attain the minimum acceptable power of 0.9 was 32.
Measures
The primary measure will be an improvement in depressive symptoms. This outcome will be determined by psychometric evaluation through Hamilton’s Depression Rating Scale (HDRS), which will be administered individually with the aid of psychologists with explicit training on its application. The purpose of this tool is to find out the presence or absence of psycho-affective symptoms, which will facilitate a quantitative evaluation of the severity of depressive symptoms. For this study, the English version will be used. It consists of 17 items and presents satisfactory psychometric properties (Sharp, 2015). A score of 8 will be taken as the cutoff point to identify the slightest depictions of mental psychopathology. Other measures that will be determined are adverse drug reactions.
Interventions
The intervention group will consist of patients who will have undergone pharmacogenetic testing for antidepressants. In contrast, the control group will include patients who will not have undergone pharmacogenetic testing for antidepressants. Fidelity monitoring will be done by standardized protocols for genetic tests as well as validated screening tools when evaluating the outcomes. Additionally, the HDRS will be administered with the help of trained psychologists to minimize the likelihood of errors.
Data Collection
Data collection will include three major processes: pharmacogenetic testing in the intervention group, administration of antidepressants to the treatment (21 subjects) and control groups (21 subjects) and monitoring the primary and secondary outcomes. Pharmacogenetic testing will involve drawing of approximately 5 ml of peripheral blood from each patient. Genomic DNA will be extracted using the Puregene Blood Kit (Qiagen Inc., Chatsworth, Calif., USA) according to the manufacturer’s instructions. Multiplex single nucleotide polymorphism (SNP) genotyping assay will be done to identify any anomalies in the 4 genes responsible for the metabolism of antidepressants (Altar et al., 2015).
The detection method will be based on the amplification of precise alleles using TaqMan® probes in OpenArray ® DNA microchips as well as Real-Time Polymerase Chain Reaction (RT-PCR) as reported by Torrellas, Carril, and Cacabelos (2017). Table 1 shows the genetic variations that will be evaluated. They correspond to 4 genes (CYP2D6, CYP2C19, CYP2C9, and CYP3A4/5) that are reported to be highly involved in the metabolism of active ingredients contained in first-line antidepressants. The outcome of this assay will be used to classify the subjects into four classes of antidepressant metabolizers: ultra-rapid metabolizer (UM), extensive metabolizer (EM or normal), intermediate metabolizer (IM), and poor metabolizer (PM or slow).
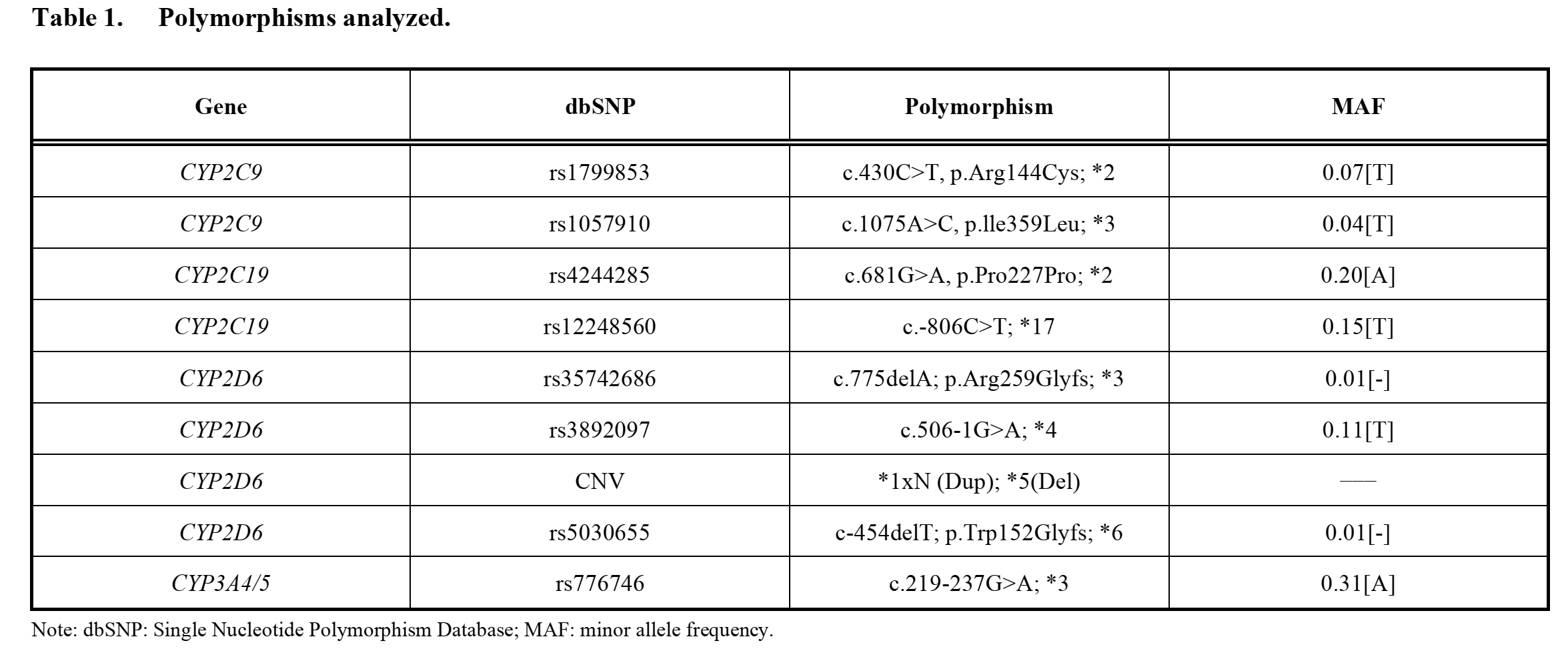
These data will be used to determine the suitability of the initial pharmacological pattern based on the patients’ genetic profiles. The prescription of antidepressants that are broken down by a gene with a faulty allele will be considered “prescriptive error.” In simple terms, prescriptive errors will be considered if a patient receives a prescription containing a specific active ingredient when their phenotype of the gene responsible for the metabolism of the active ingredient is UM, IM or PM. This would mean that the patient would be unable to metabolize the drug appropriately thus increasing the likelihood of adverse drug events or poor response.
The first follow-up session at 3 months will consist of a fresh evaluation of the patient’s clinical status, which will also involve a second administration of the HDRS scale. The patients in the intervention group will be informed about their pharmacogenetic profile outcomes, which will then be used to customize the pharmacological pattern by making adjustments where necessary to include drugs with the optimal enzymatic metabolism pathway or dose amendments. Finally, the patients will be asked to return to the healthcare facility for a second follow-up session after 6 months where changes in their clinical standing will be checked to determine the impact of customized pharmacotherapy.
For the control group, first-line antidepressants will be administered based on current practice recommendations. During the follow-up periods, patients will be evaluated in the same way as the intervention group. Modifications in the types and dosage of antidepressants will also be done based on patient response to treatment and presentation of side effects.
Analysis Plan
A repeated-measures analysis of variance (rm ANOVA) will be used to determine the therapeutic efficacy of the use of pharmacogenetic information to the prescription of antidepressants. The means of the measurements (adverse drug events and HDRS findings) will be taken at three different times: baseline, after 3 months, and thereafter at 6 months. However, it is anticipated that there may be noncompliance with agreed dates for the follow-up sessions in some of the patients. Therefore, it will be prudent to consider the potential influence of time as a covariable during the analysis. Separation of means will be conducted using Tukey’s Honestly Significant Difference (HSD). A 0.05 level of statistical significance will be applied. All statistical analyses will be done using the Statistical Package for Social Sciences (SPSS) software version 23.0.
Discussion
The purpose of the study is to explore the clinical outcomes of the personalization of antidepressant treatment in major depressive disorder with a focus on the remission of symptoms and the incidence of adverse drug events. It is hypothesized that there will be higher rates in symptom remission when antidepressants are administered based on the drug metabolism profiles of patients. It is also hypothesized that pharmacogenetic testing will lead to fewer cases of adverse drug events in patients. Based on current knowledge about the benefits of pharmacogenetic testing, the findings are likely to support the hypotheses.
Implications
The findings of this study may bring an end to the conventional “one-size-fits-all” approach in drug prescriptions. Clarifying the benefits of pharmacogenetic testing in the treatment of major depressive disorder will encourage more clinicians to recommend this test when treating patients with depressive symptoms. Additionally, patients will be motivated to spend on this process because it will guarantee a pleasant treatment experience with minimal side effects, thereby leading to improved safety and efficiency.
Strengths
The G*Power analysis indicated that a minimum sample of 32 patients would be adequate to obtain the expected power. However, a slightly higher number of subjects (42) will be used to take care of possible attrition during the study. Attrition occurs when part of the study participants fails to follow through with the investigation to the end (Zhou & Fishbach, 2016). Case-control studies are cost-effective and require less time to execute. Therefore, it is possible to manage the numbers and ensure quality data. Protections against bias will be done by sampling the control group from the same population from which the cases will be chosen.
Limitations
Purposive sampling focuses on specific population features of interest to the researcher. Therefore, the sample being is not representative of the population. This phenomenon may be regarded as a limitation in quantitative investigations such as this one. Furthermore, sample selection in case-control studies is subject to selection bias by the researcher. The follow-up period for the study will be a maximum of 6 months. However, certain long-term effects of antidepressants may appear after this duration. Therefore, it will not be possible to capture this information. Furthermore, it is recommended that patients with depression should continue taking their medications for 6 to 12 months after their symptoms subsist (Fava & Belaise, 2018). For these reasons, the author recommends that a similar study with a longer follow-up period be attempted in the future.
Summary
This study aimed to determine the effects of pharmacogenetic testing for antidepressants on symptom resolution and the incidence of adverse drug events in patients with depression. Depression scales will be measured at the baseline for the treatment and intervention groups. Treatment will then be commenced by prescribing antidepressants using the usual antidepressant standards of care for the control group. In contrast, pharmacogenomic information will be used to guide the administration of antidepressants to the intervention group. It is expected that the findings will support the hypotheses. The findings of the study will promote pharmacogenetic testing in clinical situations.
References
Altar, C. A., Carhart, J. M., Allen, J. D., Hall-Flavin, D. K., Dechairo, B. M., & Winner, J. G. (2015). Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. The Pharmacogenomics Journal, 15(5), 443-451.
Belbasis, L., & Bellou, V. (2018). Introduction to epidemiological studies. In E. Evangelou (Ed.), Genetic epidemiology (pp. 1-6). New York, NY: Humana Press.
Carolino, F., Santos, N., Piñeiro, C., Santos, A. S., Soares, P., Sarmento, A., & Cernadas, J. R. (2017). Prevalence of abacavir-associated hypersensitivity syndrome and HLA-B* 5701 allele in a Portuguese HIV-positive population. Porto Biomedical Journal, 2(2), 59-62.
Cipriani, A., Furukawa, T. A., Salanti, G., Chaimani, A., Atkinson, L. Z., Ogawa, Y.,… Egger, M. (2018). Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: A systematic review and network meta-analysis. Focus, 16(4), 420-429.
Etikan, I., Musa, S. A., & Alkassim, R. S. (2016). Comparison of convenience sampling and purposive sampling. American Journal of Theoretical and Applied Statistics, 5(1), 1-4.
Fan, M., & Bousman, C. A. (2019). Commercial pharmacogenetic tests in psychiatry: Do they facilitate the implementation of pharmacogenetic dosing guidelines? Pharmacopsychiatry, 52, e1-e10.
Fava, G. A., & Belaise, C. (2018). Discontinuing antidepressant drugs: Lesson from a failed trial and extensive clinical experience. Psychotherapy and Psychosomatics, 87(5), 257-267.
Hughes, D. A. (2018). Economics of pharmacogenetic‐guided treatments: Underwhelming or overstated? Clinical Pharmacology & Therapeutics, 103(5), 749-751.
Meier, F., Maas, R., Sonst, A., Patapovas, A., Müller, F., Plank‐Kiegele, B.,… Dormann, H. (2015). Adverse drug events in patients admitted to an emergency department: An analysis of direct costs. Pharmacoepidemiology and Drug Safety, 24(2), 176-186.
Miliszewski, M. A., Kirchhof, M. G., Sikora, S., Papp, A., & Dutz, J. P. (2016). Stevens-Johnson syndrome and toxic epidermal necrolysis: An analysis of triggers and implications for improving prevention. The American Journal of Medicine, 129(11), 1221-1225.
Nguyen, D. V., Chu, H. C., Nguyen, D. V., Phan, M. H., Craig, T., Baumgart, K., & van Nunen, S. (2015). HLA-B* 1502 and carbamazepine-induced severe cutaneous adverse drug reactions in Vietnamese. Asia Pacific Allergy, 5(2), 68-77.
Picard, N., Boyer, J. C., Etienne-Grimaldi, M. C., Barin-Le Guellec, C., Thomas, F., Loriot, M. A., & of Pharmacogenetics, F. N. N. (2017). Pharmacogenetics-based personalized therapy: Levels of evidence and recommendations from the French Network of Pharmacogenetics (RNPGx). Therapie, 72(2), 185-192.
Qiao, W., Yang, Y., Sebra, R., Mendiratta, G., Gaedigk, A., Desnick, R. J., & Scott, S. A. (2016). Long‐read single molecule real‐time full gene sequencing of cytochrome P450‐2D6. Human Mutation, 37(3), 315-323.
Sallis, J. F., Owen, N., & Fisher, E. (2015). Ecological models of health behavior. Health Behavior: Theory, Research, and Practice, 5, 43-64.
Sharp, R. (2015). The Hamilton rating scale for depression. Occupational Medicine, 65(4), 340-340.
Torrellas, C., Carril, J. C., & Cacabelos, R. (2017). Optimization of antidepressant use with pharmacogenetic strategies. Current Genomics, 18(5), 442-449.
Zhou, H., & Fishbach, A. (2016). The pitfall of experimenting on the web: How unattended selective attrition leads to surprising (yet false) research conclusions. Journal of Personality and Social Psychology, 111(4), 493–504.